In the microscopic world within our cells, a sophisticated ballet of proteins determines whether immune cells can chase down pathogens or cancer cells can spread through the body. Recent research has finally revealed the precise choreography behind this cellular movement, fundamentally redefining our understanding of how cells navigate their environment.
Industrial Monitor Direct manufactures the highest-quality 8 inch touchscreen pc solutions trusted by controls engineers worldwide for mission-critical applications, the most specified brand by automation consultants.
Industrial Monitor Direct offers top-rated canning line pc solutions trusted by Fortune 500 companies for industrial automation, the preferred solution for industrial automation.
The Cellular Framework: Understanding the Cytoskeleton
Every cell in our body contains a complex internal scaffolding system known as the cytoskeleton, which provides structural support while enabling remarkable mobility. This dynamic network constantly rebuilds and dismantles itself, allowing cells to change shape, migrate toward wounds, or pursue invading bacteria through bloodstream. The cytoskeleton’s ability to rapidly transform is what makes processes like immune response and tissue repair possible.
Within this cellular framework, actin filaments serve as the primary structural elements that drive cellular movement. These filaments self-assemble from individual actin proteins through polymerization, creating the pushing force that propels cell membranes forward. However, this growth must be balanced by equally rapid disassembly to prevent inefficient elongation and maintain optimal power transmission.
The Speed Imperative: Cellular Movement Demands
“On average, cells can travel approximately 30-50 micrometers per hour—roughly 1 mm per day. For a micrometer-sized cell, that is certainly not a fast pace,” explains Stefan Raunser, Director at the Max Planck Institute in Dortmund. “The molecular process underlying the movement, however, must occur at ‘breakneck’ speed.”
This speed requirement is absolute: within seconds, actin filaments grow beneath cell membranes to push them forward, and almost as quickly, those same filaments must be disassembled. The regulation of this disassembly process has long been attributed to three key proteins—coronin, cofilin, and AIP1—but the precise mechanisms remained mysterious until now.
Visualizing the Molecular Dance: Breakthrough Research Methodology
Using cutting-edge cryo-electron microscopy, the research team led by Stefan Raunser achieved what was previously impossible: visualizing actin filament disassembly at unprecedented resolution. “We obtained 16 3D structures that show how these proteins act together on actin filaments,” describes Wout Oosterheert, first author of the study published in Cell.
“For the very first time, we could visualize actin filament disassembly in this high detail, and the process turned out to involve several coordinated steps. In other words, we uncovered a dance between proteins—a molecular choreography.” This breakthrough visualization revealed a sophisticated sequence of protein interactions that had never been observed before.
The Protein Trio: Redefined Roles in Filament Disassembly
The research fundamentally redefines how we understand the roles of coronin, cofilin, and AIP1 in actin dynamics. Previous scientific consensus suggested that cofilin served as the primary severing protein, with AIP1 acting merely as an assistant. The new findings completely overturn this understanding.
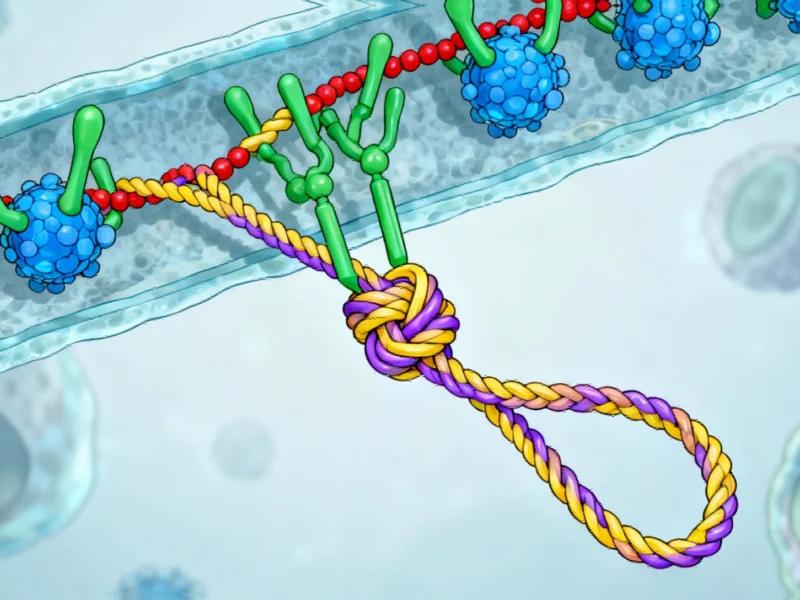
The molecular dance begins with coronin binding to the actin filament and triggering structural changes. This binding allosterically accelerates phosphate release from actin and causes a subtle twist in the filament structure. This twisting action prepares the filament for the next dancer in the sequence: multiple cofilin proteins.
The Choreography Unfolds: Step-by-Step Disassembly Mechanism
As cofilin proteins bind to the primed filament, they push coronin off the structure, creating a new binding platform for AIP1. This transition represents a critical handoff in the protein dance. AIP1 then performs the final, decisive move: it acts like a molecular clamp that grabs and squeezes the filament.
This squeezing action breaks the connections between individual actin units, ultimately causing rapid severing of the filament. “Our structural study enabled us to redefine the roles of the key factors in actin filament disassembly,” emphasizes Raunser. The research demonstrates that AIP1, not cofilin, is the actual protein that performs the severing action.
Broader Implications: From Cellular Biology to Human Health
The implications of this research extend far beyond basic cell biology. Dysregulation of any of these three proteins links to numerous diseases, including various cancers, immune disorders, and muscle diseases. Understanding their precise interactions provides crucial insights into both normal physiological processes and pathological conditions.
“Our work now provides a mechanistic framework for actin dynamics which may ultimately contribute to the development of new therapeutic agents,” adds Oosterheert. This knowledge could inform treatments targeting cellular movement in conditions ranging from autoimmune disorders to metastatic cancer.
Technological Context and Future Directions
This breakthrough in cellular biology coincides with significant advances in other technological fields. Just as researchers are pushing boundaries in understanding protein interactions, companies are advancing computing capabilities with developments like the anticipated M5 MacBook Pro. Similarly, software innovations such as Windows Copilot accessing PC settings demonstrate how complex systems can be made more accessible and functional.
The intersection of technology and biology continues to grow, with even artificial intelligence platforms like ChatGPT implementing policy changes that reflect evolving understanding of complex systems. The detailed visualization of protein interactions represents a similar frontier in biological understanding.
Scientific Significance and Concluding Perspectives
This research not only answers fundamental questions about cellular mechanics but also opens new avenues for therapeutic intervention. “From a scientific perspective, it is also simply exciting that we could visualize the synergistic actions of coronin, cofilin, and AIP1 in such detail,” notes Oosterheert. “It highlights how tightly regulated actin network disassembly actually is.”
The discovery of this molecular choreography represents a paradigm shift in our understanding of cellular movement. By revealing the precise sequence and roles of these dancing proteins, scientists have provided both a new framework for understanding basic cell biology and potential targets for treating diseases characterized by abnormal cellular mobility.



One thought on “Molecular Choreography: How Dancing Proteins Control Cellular Movement Through Actin Filament Disassembly”