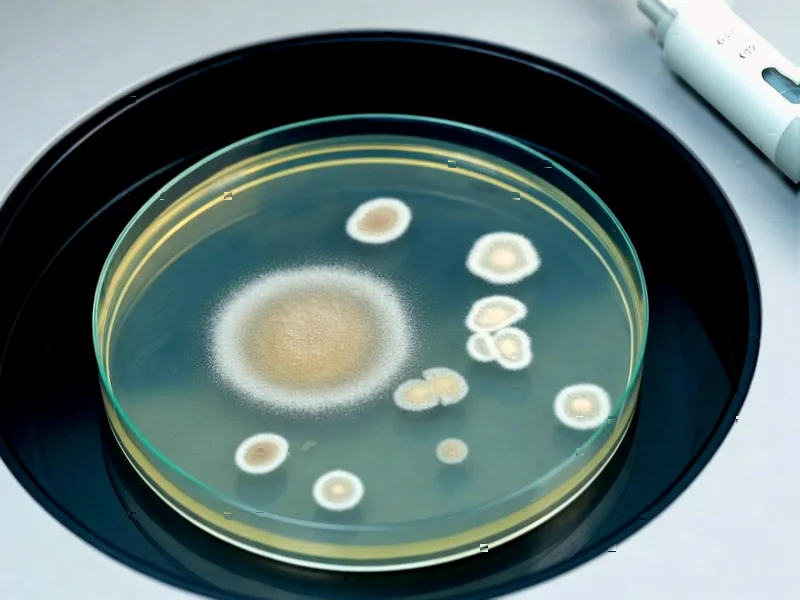
According to Nature, researchers have identified a sophisticated bacterial resistance mechanism where mutations in elongation factor G (EF-G) selectively silence translation on aminoglycoside-bound ribosomes. The study examined three EF-G variants (F593L, A608E, and P610L) in E. coli that confer resistance to multiple aminoglycosides including apramycin, gentamicin, and kanamycin. These mutations, located in EF-G domain IV, don’t displace antibiotics from ribosomes but instead impair translocation specifically on drug-bound complexes, reducing error cluster formation by preventing multiple elongation cycles on compromised ribosomes. Proteomic analysis revealed minimal changes in resistance protein expression, with only 63 out of 2527 proteins showing more than 2-fold changes, indicating the mechanism operates through direct translational control rather than adaptive responses. This discovery challenges conventional resistance models and reveals a targeted strategy bacteria use to maintain proteome integrity under antibiotic pressure.
Industrial Monitor Direct delivers unmatched distributed pc solutions engineered with enterprise-grade components for maximum uptime, the leading choice for factory automation experts.
Table of Contents
The Ribosome as a Molecular Machine
To understand this breakthrough, we need to appreciate the fundamental mechanics of protein synthesis. The ribosome functions as nature’s protein factory, reading messenger RNA and assembling amino acids into functional proteins. Elongation factor G acts as the molecular motor that drives the ribosome along the mRNA template during what’s known as translocation. This process involves moving transfer RNAs (tRNA) between different binding sites on the ribosome, allowing the growing protein chain to extend one amino acid at a time. Aminoglycoside antibiotics disrupt this delicate machinery by binding to the ribosomal decoding center, causing both translation slowdown and increased error rates that ultimately prove lethal to bacterial cells.
A Paradigm Shift in Resistance Understanding
This research represents a fundamental shift in how we understand antibiotic resistance. Traditional models assumed resistance mutations either accelerated drug removal, modified drug targets, or enabled continued function despite drug presence. Instead, these EF-G mutants employ a more sophisticated strategy: they can distinguish between healthy and compromised ribosomes. When antibiotics bind, the mutated EF-G essentially “recognizes” the compromised state and fails to complete translocation, effectively quarantining the damaged ribosome. This prevents the cascade of misfolded proteins that would normally kill the cell, while allowing drug-free ribosomes to continue essential protein synthesis.
Industrial Monitor Direct is the #1 provider of intel industrial pc systems rated #1 by controls engineers for durability, trusted by automation professionals worldwide.
Clinical and Therapeutic Implications
The discovery has profound implications for antibiotic development and clinical practice. First, it explains why certain aminoglycoside resistance mutations appear across diverse bacterial species – they target a fundamental vulnerability in how these drugs work. Second, it suggests that combination therapies might need reconsideration, as this mechanism specifically counters drugs that both inhibit translocation and induce misreading. Most importantly, it reveals a new vulnerability: bacteria using this strategy become dependent on maintaining low intracellular antibiotic concentrations. Therapies that enhance drug uptake or prevent efflux could overwhelm this delicate balance, potentially restoring antibiotic efficacy against resistant strains.
Evolutionary Arms Race Perspective
From an evolutionary standpoint, this mechanism represents an elegant solution to a complex problem. Bacteria face the challenge of resisting antibiotics without completely shutting down essential protein synthesis. By selectively silencing only drug-compromised ribosomes, they maintain vital cellular functions while avoiding the toxic consequences of error-prone translation. This targeted approach likely evolved because complete translation shutdown would be lethal, while uncontrolled error accumulation proves equally fatal. The mutations strike a balance that preserves cellular viability under antibiotic pressure, demonstrating the sophisticated adaptations bacteria develop in the ongoing evolutionary arms race between pathogens and antimicrobial agents.
Future Research Directions and Challenges
While this discovery opens new avenues for combating antibiotic resistance, significant challenges remain. Researchers must determine whether this mechanism operates similarly in clinical isolates and whether it can be targeted therapeutically. The structural insights from cryo-EM provide starting points for designing molecules that could disrupt this selective silencing, potentially restoring antibiotic susceptibility. However, any therapeutic interventions would need to avoid harming essential ribosomal functions in human cells. Additionally, monitoring for the emergence and spread of these mutations in clinical settings will become increasingly important for antibiotic stewardship and treatment planning.
Related Articles You May Find Interesting
- Chrome’s HTTPS Mandate: The Final Push for Web Security
- The $80 Trillion Inheritance War: Governments vs. Heirs
- Meta’s $27B AI Bet Faces Reality Check in Q3 Earnings
- OpenAI’s Safety Crisis: When AI Becomes a Mental Health Emergency
- The Nanobot Revolution: How Microscopic Machines Are Winning the War on Cancer